Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
Micro-computed tomography (μCT) is a valuable imaging modality for longitudinal quantification of bone volumes to identify disease or treatment effects for a broad range of conditions that affect bone health. Complex structures, such as the hindpaw with up to 31 distinct bones in mice, have considerable analytic potential, but quantification is often limited to a single bone volume metric due to the intensive effort of manual segmentation. Herein, we introduce a high-throughput, user-friendly, and semi-automated method for segmentation of murine hindpaw μCT datasets.
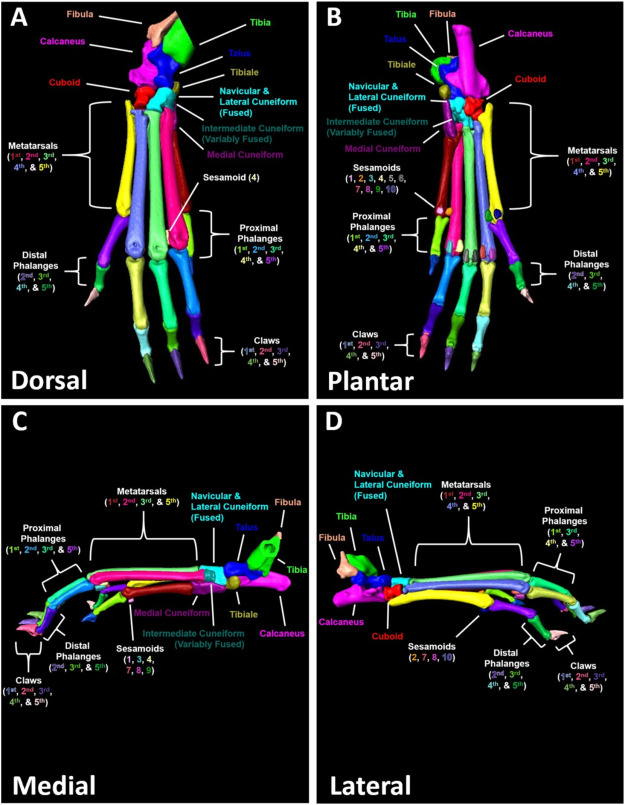
In vivo μCT was performed on male (n = 4; 2–8-months) and female (n = 4; 2–5-months) C57BL/6 mice longitudinally each month. Additional 9.5-month-old male C57BL/6 hindpaws (n = 6 hindpaws) were imaged by ex vivo μCT to investigate the effects of resolution and integration time on analysis outcomes. The DICOMs were exported to Amira software for the watershed-based segmentation, and watershed markers were generated automatically at approximately 80% accuracy before user correction. The semi-automated segmentation method utilizes the original data, binary mask, and bone-specific markers that expand to the full volume of the bone using watershed algorithms.
Compared to the conventional manual segmentation using Scanco software, the semi-automated approach produced similar raw bone volumes. The semi-automated segmentation also demonstrated a significant reduction in segmentation time for both experienced and novice users compared to standard manual segmentation. ICCs between experienced and novice users were >0.9 (excellent reliability) for all but 4 bones.
The described semi-automated segmentation approach provides remarkable reliability and throughput advantages. Adoption of the semi-automated segmentation approach will provide standardization and reliability of bone volume measures across experienced and novice users and between institutions. The application of this model provides a considerable strategic advantage to accelerate various research opportunities in pre-clinical bone and joint analysis towards clinical translation.
“The DICOMs were exported to Amira software for the watershed-based segmentation, and watershed markers were generated automatically at approximately 80% accuracy before user correction. The semi-automated segmentation method utilizes the original data, binary mask, and bone-specific markers that expand to the full volume of the bone using watershed algorithms. Herein, we developed a high-throughput and user-friendly semi-automated segmentation workflow for murine hindpaw μCT datasets using commercially available algorithms in Amira software. As one of the major roadblocks to widespread adoption of automated segmentation approaches is the intensive computer-based knowledge necessary for application, we validated that the segmentation strategy could be quickly adopted with minimal training by novice users with no prior experience using the Amira software. The described segmentation method demonstrated significant benefits in throughput compared to conventional μCT analysis performed by manual contouring, and the approach also showed excellent internal consistency and inter-user reliability. Thus, we have established an automated approach to isolate individual small bones in complex structures, such as the hindpaw, for μCT analysis with the capacity for widespread adoption.”
For Research Use Only. Not for use in diagnostic procedures.