Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
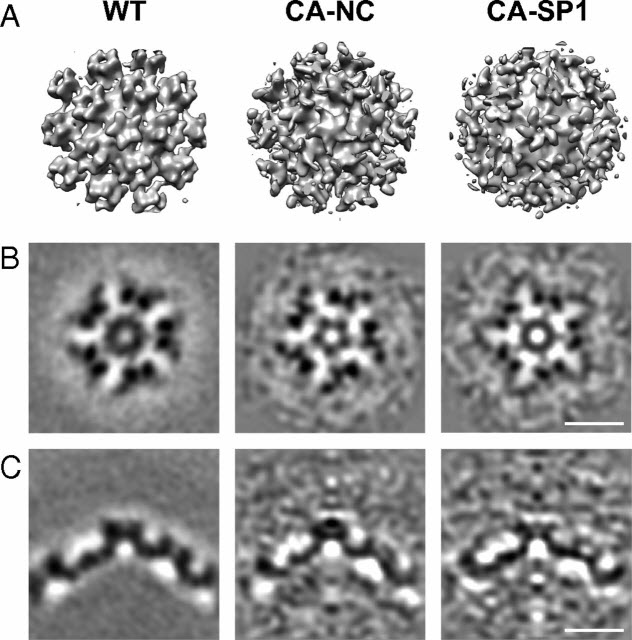
HIV-1 maturation occurs via multiple proteolytic cleavages of the Gag polyprotein, causing rearrangement of the virus particle required for infectivity. (…) How individual cleavages contribute to changes in protein structure and interactions, and how the mature, conical capsid forms, are poorly understood. Here, we employed cryoelectron tomography to determine morphology and high-resolution CA lattice structures for HIV1 derivatives in which Gag cleavage sites are mutated. These analyses prompt us to revise current models for the crucial maturation switch.
The approximate outlines of complete and incomplete mature cores were traced on X–Y slices through 8x binned tomograms of CA-NC viruses, using the segmentation function of Amira (versions 5.5 and 6.1.1). These paths were interpolated in Amira to produce a binary volume mask for each core, which was filtered with a circular Gaussian kernel of 20 pixels in MATLAB using the tom_filter function of the TOM package and utilized to generate isosurfaces. Points with a spacing of two pixels were sampled along the isosurfaces at an intensity threshold of 0.3 and were assigned Euler angles based on their orientation relative to the normal of the surface at each point.
For Research Use Only. Not for use in diagnostic procedures.