Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
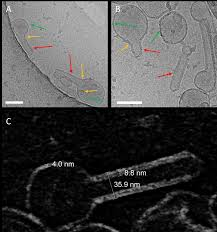
The matrix protein M1 of the Influenza A virus is considered to mediate viral assembly and budding at the plasma membrane (PM) of infected cells. In order for a new viral particle to form, the PM lipid bilayer has to bend into a vesicle towards the extracellular side. Studies in cellular models have proposed that different viral proteins might be responsible for inducing membrane curvature in this context (including M1), but a clear consensus has not been reached. In this study, we use a combination of fluorescence microscopy, cryogenic transmission electron microscopy (cryo-TEM), cryo-electron tomography (cryo-ET) and scanning fluorescence correlation spectroscopy (sFCS) to investigate M1-induced membrane deformation in biophysical models of the PM. Our results indicate that M1 is indeed capable to cause membrane curvature in lipid bilayers containing negatively-charged lipids, in the absence of other viral components. Furthermore, we prove that simple protein binding is not sufficient to induce membrane restructuring. Rather, it appears that stable M1-M1 interactions and multimer formation are required in order to alter the bilayer three-dimensional structure, through the formation of a protein scaffold. Finally, our results suggest that, in a physiological context, M1-induced membrane deformation might be modulated by the initial bilayer curvature and the lateral organization of membrane components (i.e. the presence of lipid domains).
Amira Software was used for visualization.
For Research Use Only. Not for use in diagnostic procedures.