Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
Our study placed emphasis on solving problems in processing high-throughput bright field images and made attempt in developing a method for the extraction and reconstruction of multiple structures. This will facilitate a better understanding of the cerebral anatomical features under the pathological state of AD and shows extensive application prospect in drug efficacy assessment from brain-wide level.
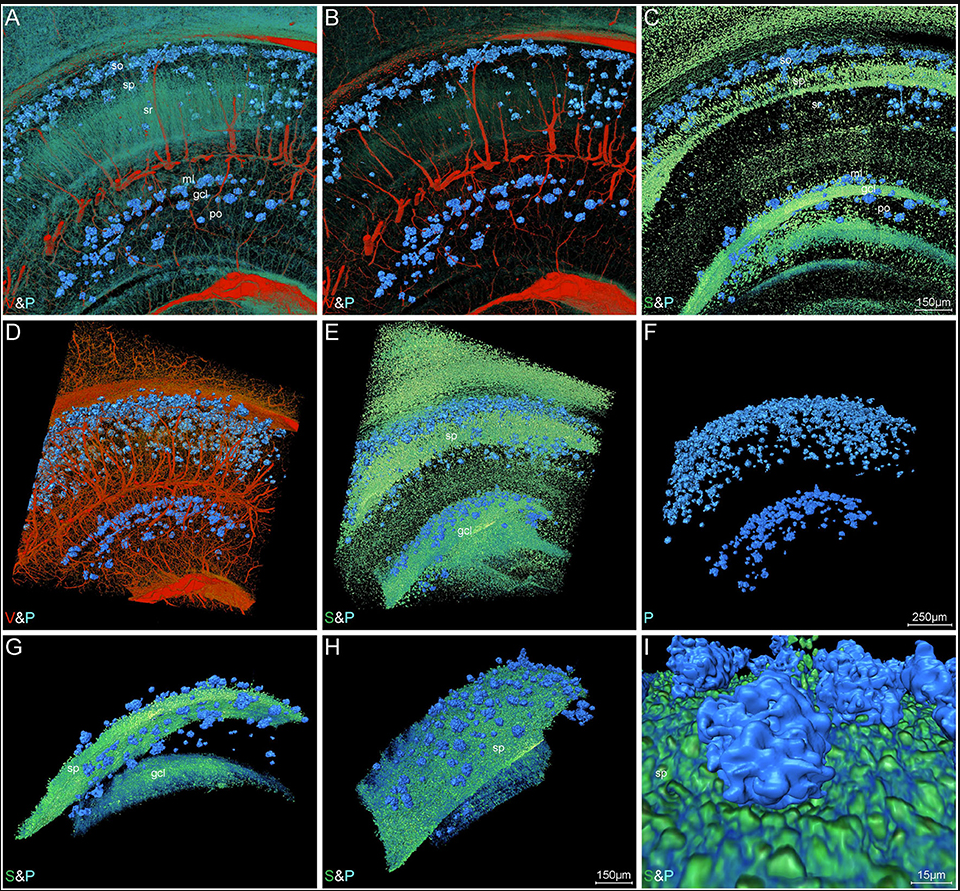
Simultaneously visualizing Amyloid-β (Aβ) plaque with its surrounding brain structures at the subcellular level in the intact brain is essential for understanding the complex pathology of Alzheimer’s disease, but is still rarely achieved due to the technical limitations. Combining the micro-optical sectioning tomography (MOST) system, whole-brain Nissl staining, and customized image processing workflow, we generated a whole-brain panorama of Alzheimer’s disease mice without specific labeling. The workflow employed the steps that include virtual channel splitting, feature enhancement, iso-surface rendering, direct volume rendering, and feature fusion to extract and reconstruct the different signals with distinct gray values and morphologies. Taking advantage of this workflow, we found that the denser-distribution areas of Aβ plaques appeared with relatively more somata and smaller vessels, but show a dissimilar distributing pattern with nerve tracts. In addition, the entorhinal cortex and adjacent subiculum regions present the highest density and biggest diameter of plaques. The neuronal processes in the vicinity of these Aβ plaques showed significant structural alternation such as bending or abrupt branch ending. The capillaries inside or adjacent to the plaques were observed with abundant distorted micro-vessels and abrupt ending. Depicting Aβ plaques, somata, nerve processes and tracts, and blood vessels simultaneously, this panorama enables us for the first time, to analyze how the Aβ plaques interact with capillaries, somata, and processes at a submicron resolution of 3D whole-brain scale, which reveals potential pathological effects of Aβ plaques from a new cross-scale view. Our approach opens a door to routine systematic studies of complex interactions among brain components in mouse models of Alzheimer’s disease.
The code includes scripting commands using Tool Command Language (Tcl) with Amira-specific extensions. It allows users to automate certain processes and to create scripts for managing routine tasks or for presenting demos in the Amira software (Amira 2019.2). In brief, before executing the Tcl code, the data directory path must be set in the scripts. The location where should input the data directory path had been annotated in the word file “Scripts for Virtual channel splitting.” Then, copy all the scripts to a TXT file and change the file extension as “.hx.” Next, drag and drop the “.hx” file onto the Amira console to run the Tcl code in the Amira software. […] Iso-surface rendering and direct volume rendering were employed to achieve high-quality rendering of brain structures using Amira software. […] The somata, nerve processes, and tracts were reconstructed using direct volume rendering in Amira software.
For Research Use Only. Not for use in diagnostic procedures.