Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
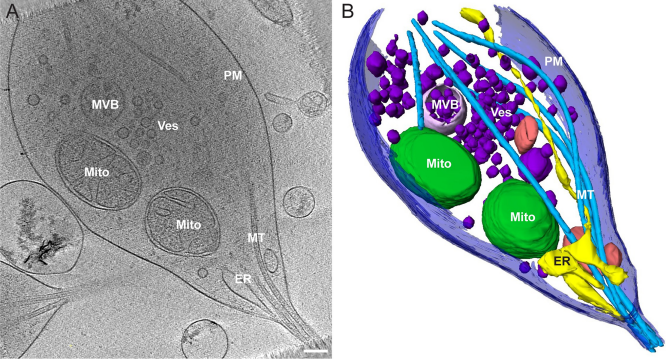
Neurons project axons to local and distal sites and can display heterogeneous morphologies with limited physical dimensions that may influence the structure of large organelles such as mitochondria. Using cryo-electron tomography (cryo-ET), we characterized native environments within axons and presynaptic varicosities to examine whether spatial restrictions within these compartments influence the morphology of mitochondria. Segmented tomographic reconstructions revealed distinctive morphological characteristics of mitochondria residing at the narrowed boundary between presynaptic varicosities and axons with limited physical dimensions (approximately 80 nm), compared to mitochondria in nonspatially restricted environments. Furthermore, segmentation of the tomograms revealed discrete organizations between the inner and outer membranes, suggesting possible independent remodeling of each membrane in mitochondria at spatially restricted axonal/varicosity boundaries. Thus, cryo-ET of mitochondria within axonal subcompartments reveals that spatial restrictions do not obstruct mitochondria from residing within them, but limited available space can influence their gross morphology and the organization of the inner and outer membranes. These findings offer new perspectives on the influence of physical and spatial characteristics of cellular environments on mitochondrial morphology and highlight the potential for remarkable structural plasticity of mitochondria to adapt to spatial restrictions within axons.
Reconstructed tomograms were further processed using the median, nonlocal means and Lanczos filters in Amira for manual and semiautomated segmentation. Segmentation was accomplished by manually tracing membranes for each Z slice of the tomographic data set. Membranes were identified and segmented with reference to visualization in all 3 dimensions (X, Y, and Z). The brush tool was primarily used in manual segmentation. When possible, masking approaches were also used in combination with density thresholding for semiautomated segmentation. After segmentation, smoothing tools were employed for the manual tracings, and surfaces were rendered for model construction. All measurements (length and diameter) were performed in either Amira using the 3D measurement tool or in IMOD.
For Research Use Only. Not for use in diagnostic procedures.