Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
Precise methods for quantifying drug accumulation in brain tissue are currently very limited, challenging the development of new therapeutics for brain disorders. Transcardial perfusion is instrumental for removing the intravascular fraction of an injected compound, thereby allowing for ex vivo assessment of extravasation into the brain. However, pathological remodeling of tissue microenvironment can affect the efficiency of transcardial perfusion, which has been largely overlooked.
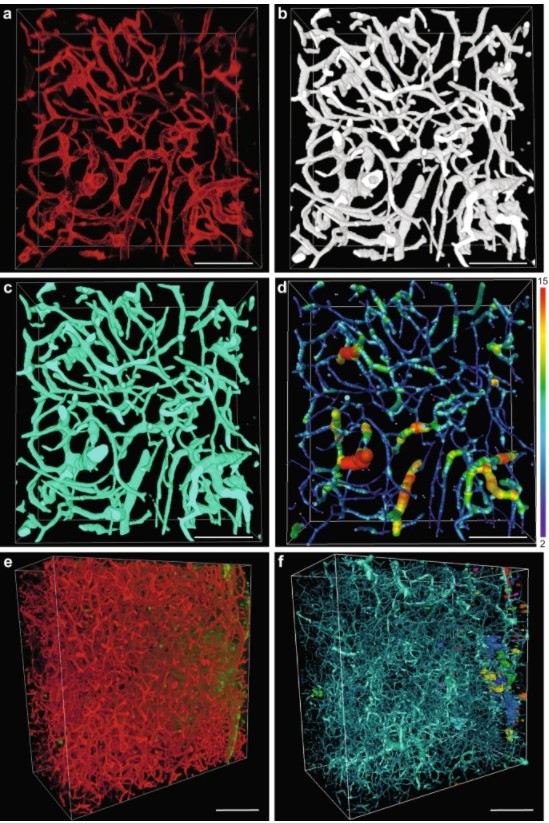
We show that, in contrast to healthy vasculature, transcardial perfusion cannot remove an injected compound from the tumor vasculature to a sufficient extent leading to considerable overestimation of compound extravasation. We demonstrate that 3D deep imaging of optically cleared tumor samples overcomes this limitation. We developed two machine learning-based semi-automated image analysis workflows, which provide detailed quantitative characterization of compound extravasation patterns as well as tumor angioarchitecture in large three-dimensional datasets from optically cleared samples. This methodology provides a precise and comprehensive analysis of extravasation in brain tumors and allows for correlation of extravasation patterns with specific features of the heterogeneous brain tumor vasculature.
Afterward, images were imported to Amira and extravasation spots were recognized and measured using Label analysis module. Spots smaller than 10,000 µm3 were excluded from the analysis. For method validation, two image subsets 2 mm3 in volume were selected from two different tumors, number of extravasation spots was counted manually by human annotator and by using developed workflow. Postprocessed datasets were imported to Amira and the vasculature was traced by the AutoSkeleton module employing method developed by Fouard et al.
For Research Use Only. Not for use in diagnostic procedures.