Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
Adipose models have been applied to mechanistic studies of metabolic diseases (such as diabetes) and the subsequent discovery of new therapeutics. However, typical models are either insufficiently complex (2D cell cultures) or expensive and labor intensive (mice/in vivo). To bridge the gap between these models and in order to better inform pre-clinical studies we have developed a drug-responsive 3D model of white adipose tissue (WAT).
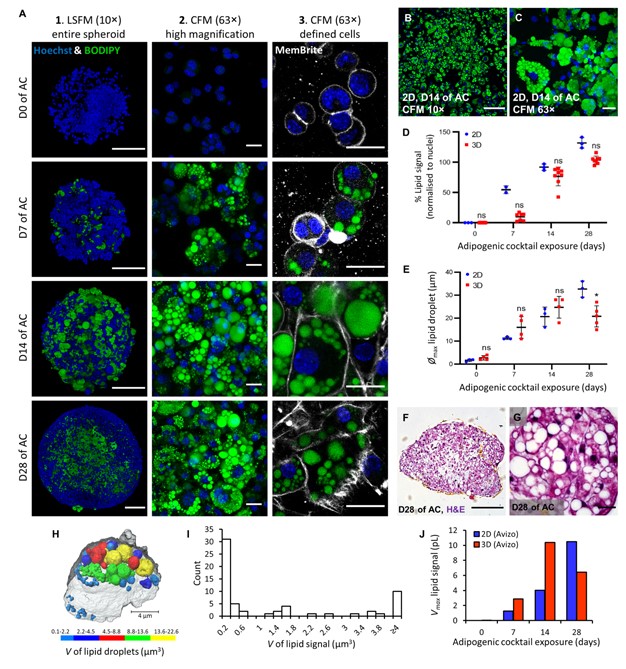
Here, spheroids (680 ± 60 μm) comprising adipogenic 3T3-L1 cells encapsulated in 3D matrix were fabricated manually on a 96 well scale. Spheroids were highly characterised for lipid morphology, selected metabolite and adipokine secretion, and gene expression; displaying significant upregulation of certain adipogenic-specific genes compared with a 2D model. Furthermore, induction of lipolysis and promotion of lipogenesis in spheroids could be triggered by exposure to 8-br-cAMP and oleic-acid respectively. Metabolic and high content imaging data of spheroids exposed to an adipose-targeting drug, rosiglitazone, resulted in dose-responsive behavior. Thus, our 3D WAT model has potential as a powerful scalable tool for compound screening and for investigating adipose biology.
The image processing was performed using Thermo Scientific Amira-Avizo 9.5 software. The 3D dataset were filtered using a non-local means algorithm to reduce noise (denoise) the images while preserving the boundaries between the lipid droplets within the spheroid cells. The nuclei and lipid droplets were segmented using an interactive thresholding, which enabled computing of their respective volumes for each time point. For an image set of a day of AC exposure spheroid, a few selected cells were segmented using a gradient-based watershed algorithm and used to mask the intracellular lipid droplets. A distance transform watershed algorithm was applied to separate overlapping lipid droplets. All the droplets were individually labelled and a label analysis was performed to compute their volumes. The lipid droplets were finally sieved in five regions and visualised in 3D. These values are then ranked to construct the volume distribution with a class interval size of 0.2 μm.
For Research Use Only. Not for use in diagnostic procedures.