Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
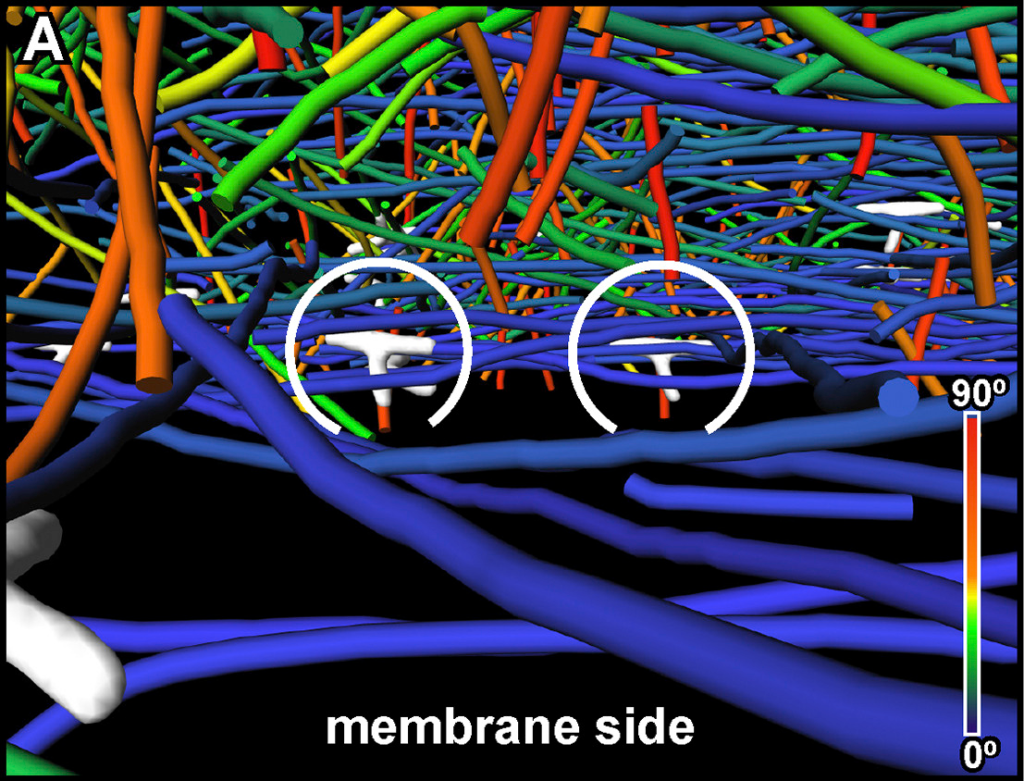
Actin waves are dynamic supramolecular structures involved in cell migration, cytokinesis, adhesion, and neurogenesis. Although wave-like propagation of actin networks is a widespread phenomenon, the actin architecture underlying wave propagation remained unknown. In situ cryo-electron tomography of Dictyostelium cells unveils the wave architecture and provides evidence for wave progression by de novo actin nucleation. Subtomogram averaging reveals the structure of Arp2/3 complex-mediated branch junctions in their native state, and enables quantitative analysis of the 3D organization of branching within the waves.
The 3D reconstruction with a final pixel size of 1.684 nm was obtained by weighted-back projection using IMOD and rendered in 3D using Amira. Filaments were automatically segmented using the actin segmentation package in Amira.[…] Binned twice tomograms were subjected to nonlocal-means filtering using Amira software provided by Thermo Fisher Scientific. Actin filaments were traced by an automated segmentation algorithm based on a generic filament as a template, with a diameter and length of 8 and 42 nm, respectively. To reduce background noise, short filamentous structures with lengths below 70 nm (or 40 nm in Figure 7) were filtered out.
For Research Use Only. Not for use in diagnostic procedures.