Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
For decades, clearing and staining with Alcian Blue and Alizarin Red has been the gold standard to image vertebrate skeletal development. Here, we present an alternate approach to visualise bone and cartilage based on X-ray microCT imaging, which allows the collection of genuine 3D data of the entire developing skeleton at micron resolution.
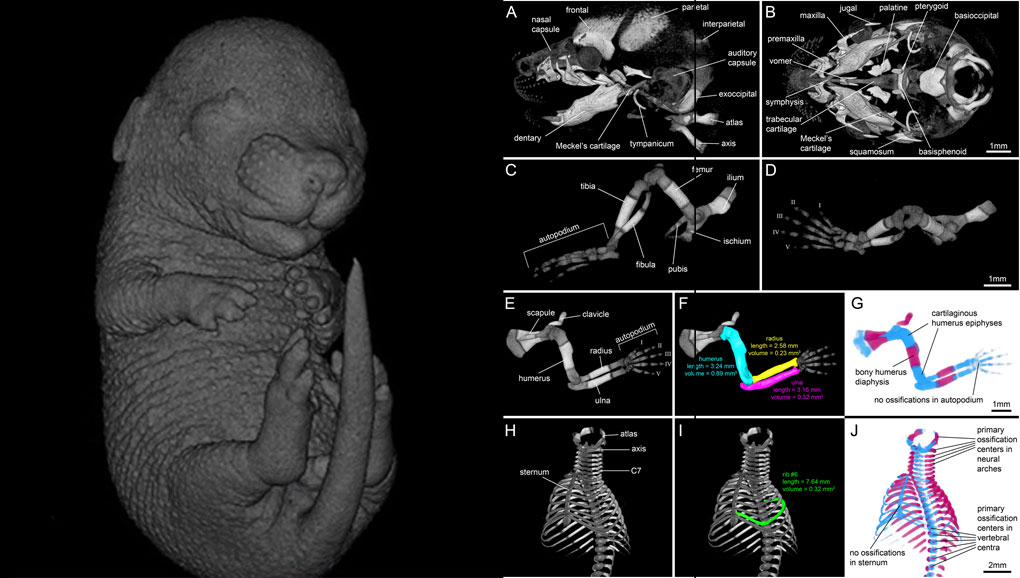
Our novel protocol is based on ethanol fixation and staining with Ruthenium Red, and efficiently contrasts cartilage matrix, as demonstrated in whole E16.5 mouse foetuses and limbs of E14 chicken embryos. Bone mineral is well preserved during staining, thus the entire embryonic skeleton can be imaged at high contrast. Differences in X-ray attenuation of ruthenium and calcium enable the spectral separation of cartilage matrix and bone by dual energy microCT (microDECT). Clearing of specimens is not required. The protocol is simple and reproducible. We demonstrate that cartilage contrast in E16.5 mouse foetuses is adequate for fast visual phenotyping. Morphometric skeletal parameters are easily extracted. We consider the presented workflow to be a powerful and versatile extension to the toolkit currently available for qualitative and quantitative phenotyping of vertebrate skeletal development.
MicroCT scans were imported into the commercial 3D reconstruction software package Amira 2019.1 . Volumes were filtered with a 3D bilateral filter and a 3D Gaussian filter to increase signal-to-noise ratio. Image intensities were standardised to Hounsfield units (HU) based on measurement of a water-air phantom. For visualization of the staining results in the 60 kVp scans of mouse foetuses we used virtual slices and volume renderings using different histogram settings in analogy to windowing in clinical computed tomography (soft tissue window, 100-400 HU; cartilage window, 400-900 HU; bone window, 900-2000 HU). For displaying selected parts of the foetal mouse skeleton, such as the skull, forelimb, hind limb and ribcage, we used the Amira volume segmentation tool Volume edit to crop selected body regions.
For Research Use Only. Not for use in diagnostic procedures.