Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
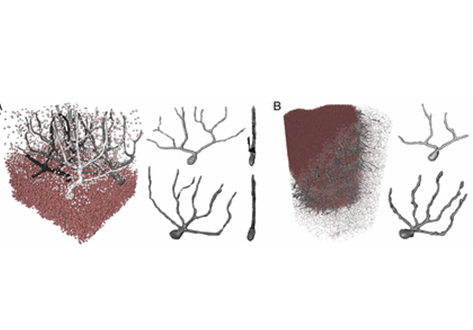
To quantitatively evaluate brain tissue and its corresponding function, knowledge of the 3D cellular distribution is essential. The gold standard to obtain this information is histology, a destructive and labor-intensive technique where the specimen is sliced and examined under a light microscope, providing 3D information at nonisotropic resolution. To overcome the limitations of conventional histology, we use phase-contrast X-ray tomography with optimized optics, reconstruction, and image analysis, both at a dedicated synchrotron radiation endstation, which we have equipped with X-ray waveguide optics for coherence and wavefront filtering, and at a compact laboratory source. As a proof-of-concept demonstration we probe the 3D cytoarchitecture in millimeter-sized punches of unstained human cerebellum embedded in paraffin and show that isotropic subcellular resolution can be reached at both setups throughout the specimen. To enable a quantitative analysis of the reconstructed data, we demonstrate automatic cell segmentation and localization of over 1 million neurons within the cerebellar cortex. This allows for the analysis of the spatial organization and correlation of cells in all dimensions by borrowing concepts from condensed-matter physics, indicating a strong short-range order and local clustering of the cells in the granular layer. By quantification of 3D neuronal “packing,” we can hence shed light on how the human cerebellum accommodates 80% of the total neurons in the brain in only 10% of its volume. In addition, we show that the distribution of neighboring neurons in the granular layer is anisotropic with respect to the Purkinje cell dendrites.
The 3D visualization of the reconstructed data was carried out with Avizo Lite 9 (FEI Visualization Sciences Group).
For Research Use Only. Not for use in diagnostic procedures.