Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
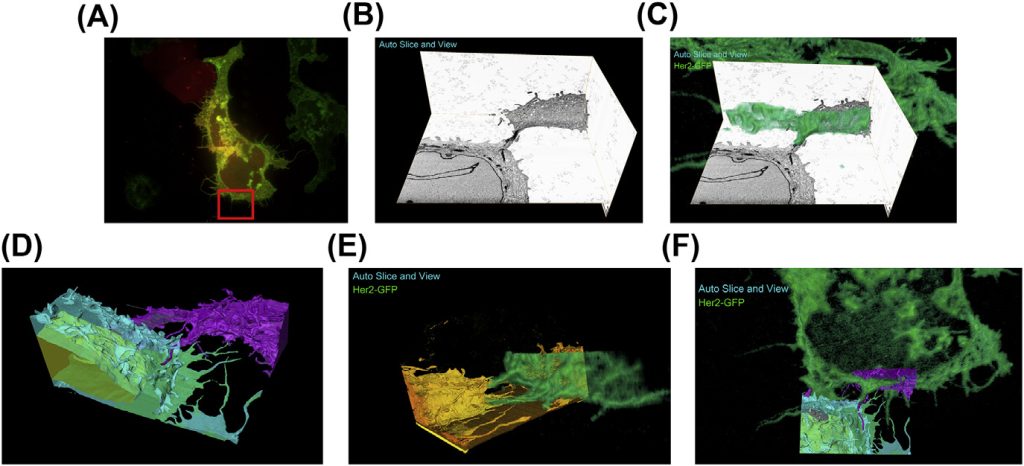
While fluorescence microscopy provides tools for highly specific labeling and sensitive detection, its resolution limit and lack of general contrast has hindered studies of cellular structure and protein localization. Recent advances in correlative light and electron microscopy (CLEM), including the fully integrated CLEM workflow instrument, the Thermo Scientific CorrSight with MAPS, have allowed for a more reliable, reproducible, and quicker approach to correlate three-dimensional time-lapse confocal fluorescence data, with three-dimensional focused ion beam–scanning electron microscopy data. Here we demonstrate the entire integrated CLEM workflow using fluorescently tagged MCF7 breast cancer cells.
For Research Use Only. Not for use in diagnostic procedures.