Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
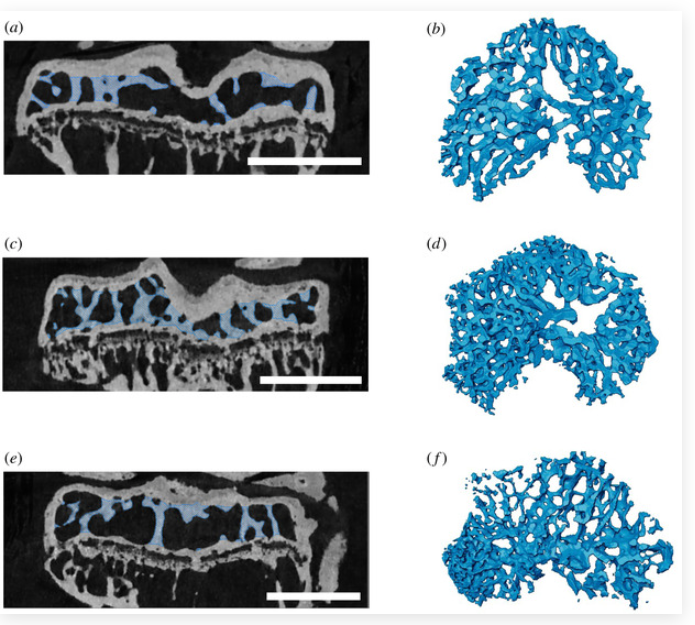
Many physiological, biomechanical, evolutionary and clinical studies that explore skeletal structure and function require successful separation of trabecular from cortical compartments of a bone that has been imaged by X-ray micro-computed tomography (micro-CT) prior to analysis. Separation often involves manual subdivision of these two similarly radio-opaque compartments, which can be time-consuming and subjective. We have developed an objective, semi-automated protocol which reduces user bias and enables straightforward, user-friendly segmentation of trabecular from the cortical bone without requiring sophisticated programming expertise. This method can conveniently be used as a ‘recipe’ in commercial programmes (Avizo herein) and applied to a variety of datasets. Here, we characterize and share this recipe, and demonstrate its application to a range of murine and human bone types, including normal and osteoarthritic specimens, and bones with distinct embryonic origins and spanning a range of ages. We validate the method by testing inter-user bias during the scan preparation steps and confirm utility in the architecturally challenging analysis of growing murine epiphyses. We also report details of the recipe, so that other groups can readily re-create a similar method in open access programmes. Our aim is that this method will be adopted widely to create a reproducible and time-efficient method of segmenting trabecular and cortical bone.
Our method relies on an input segmentation of the marrow space and can easily be loaded as a ‘recipe’ in Avizo, which enables all further steps to run automatically. We compare this to other automatic segmentation methods to build upon previous advances. […], This preprocessing step and algorithm implementation was performed in Avizo (Thermo-Scientific, v. 2019.2 and 2020.1). […], We preprocessed the dataset in Avizo, although other segmentation software could also be used. First, we filtered the CT scans using a non-local means filter (three-dimensional GPU adaptive manifold setting) to remove noise in the scans and facilitate the watershed operation (figure 1a). Next, we separated the bone of interest from the rest of the scan by placing ‘seeds’ and assigning them to two different regions (defined as ‘materials’ in Avizo): the first containing both the bone of interest and its marrow space, and the second being the background (any bone(s) not included in analysis and background voxels) (figure 1b). […], Avizo is one of the most commonly used segmentation programmes by researchers and creating an Avizo ‘recipe’ for this method makes it very easy to implement for Avizo users in particular. Loading our recipe in Avizo does not require any coding knowledge; after downloading the ‘recipe’, the user selects the preprocessed marrow space as the recipe input and all steps will be run in sequence. In addition to making the Avizo ‘recipe’ freely available, we share its specific steps in a Github repository (https://github.com/evaherbst/Trabecular_Segmentation_Avizo) so that the method can also be applied in open-source programmes which are more accessible and may be preferred by other researchers. etc..
For Research Use Only. Not for use in diagnostic procedures.