Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
The Braconidae are a megadiverse and ecologically highly important group of insects. The vast majority of braconid wasps are parasitoids of other insects, usually attacking the egg or larval stages of their hosts. The ovipositor plays a crucial role in the assessment of the potential host and precise egg laying. We used lightand electron-microscopic techniques to investigate all inherent cuticular elements of the ovipositor (the female 9th abdominal tergum, two pairs of valvifers, and three pairs of valvulae) of the braconid Habrobracon hebetor (Say, 1836) in detail with respect to their morphological structure and microsculpture.
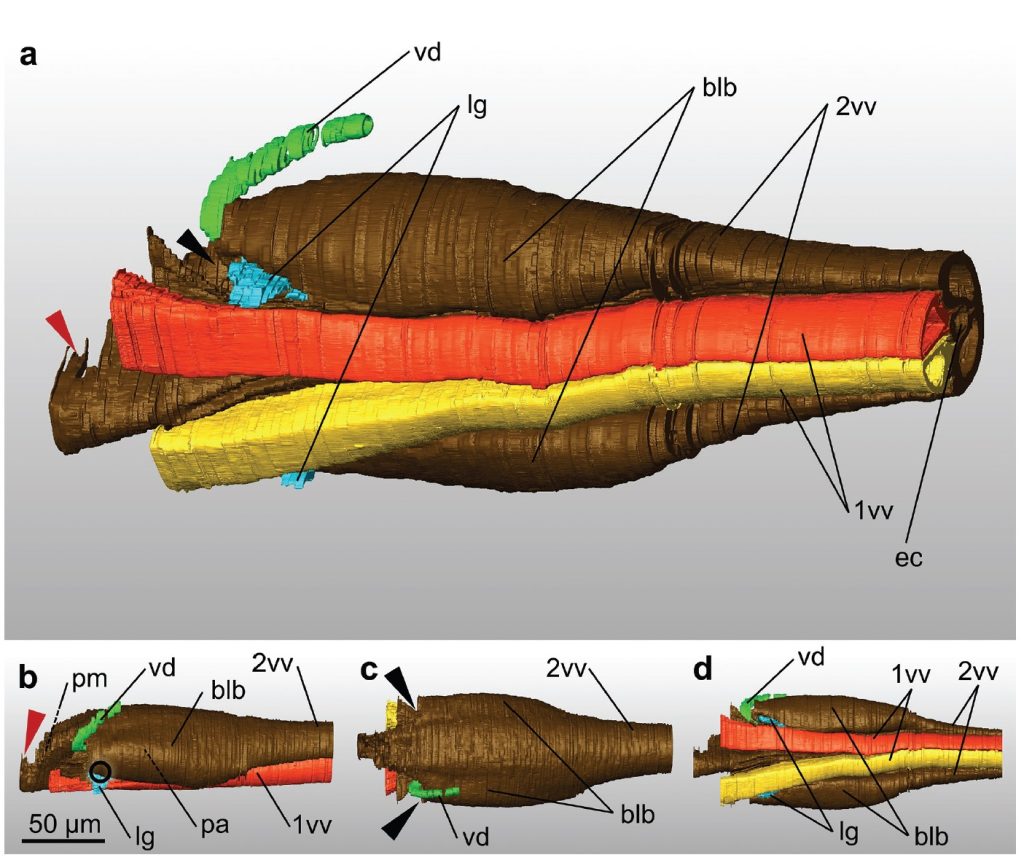
Based on serial sections, we reconstructed the terebra in 3D with all its inherent structures and the ligaments connecting it to the 2nd valvifers. We examined the exact position of the paired valvilli, which are bilateral concave structures that protrude into the egg canal. In H. hebetor, these structures putatively divert the egg ventrally between the 1st valvulae for oviposition. We discuss further mechanical and functional aspects of the ovipositor in order to increase the understanding of this putative key feature in the evolution of braconids and of parasitoid wasps in general.
The aligned image stack was then imported into Amira version 6.0. We pre-segmented the 1st and 2nd valvulae, the duct of the venom gland and the ligaments that connect the terebra and the 2nd valvifer in the software’s segmentation editor by manually labeling approximately every 15th virtual slice along the terebra and every 4th virtual slice in the proximal bulbous region and assigned them to different ‘materials’. The labels served as an input for automated segmentation by using the Biomedical Image Segmentation App Biomedisa (available online at https://biomedisa.de) (Lösel et al. 2020). The output of Biomedisa was then partially corrected manually in Amira, and a final surface model was generated.
For Research Use Only. Not for use in diagnostic procedures.