Thermo Fisher Scientific › Electron Microscopy › Electron Microscopes › 3D Visualization, Analysis and EM Software › Use Case Gallery
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the COVID19 pandemic, is a highly pathogenic β-coronavirus. As other coronaviruses, SARS-CoV-2 is enveloped, replicates in the cytoplasm and assembles at intracellular membranes. Here, we structurally characterize the viral replication compartment and report critical insights into the budding mechanism of the virus, and the structure of extracellular virions close to their native state by in situ cryo-electron tomography and subtomogram averaging.
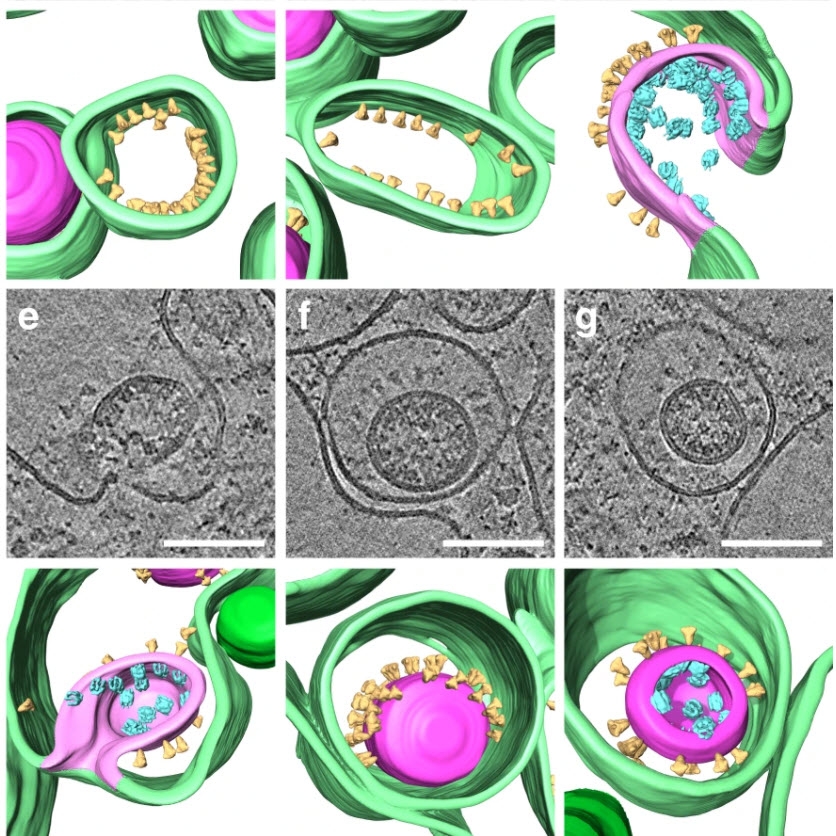
We directly visualize RNA filaments inside the double membrane vesicles, compartments associated with viral replication. The RNA filaments show a diameter consistent with double-stranded RNA and frequent branching likely representing RNA secondary structures. We report that assembled S trimers in lumenal cisternae do not alone induce membrane bending but laterally reorganize on the envelope during virion assembly. The viral ribonucleoprotein complexes (vRNPs) are accumulated at the curved membrane characteristic for budding sites suggesting that vRNP recruitment is enhanced by membrane curvature. Subtomogram averaging shows that vRNPs are distinct cylindrical assemblies. We propose that the genome is packaged around multiple separate vRNP complexes, thereby allowing incorporation of the unusually large coronavirus genome into the virion while maintaining high steric flexibility between the vRNPs. Here, we used cryo-electron tomography (cryo-ET) on cryo-focused ion beam milled lamellae or whole-cell cryo-ET of various SARS-CoV-2-infected cell lines, which were chemically fixed for biosafety reasons, to structurally characterize DMV morphology, virus assembly, and extracellular virions close to their native state.
(…) denoised tomogram was further processed with Amira 2019.3 (Thermo Fisher Scientific) using the Membrane Enhancement Filter with a feature scale of 6.5 nm. An initial segmentation was created using the Top-hat tool. This initial segmentation was manually refined by removing segmented sections derived from noise and by manually adding segmentations of membranes where automated segmentation failed. [RNA] Tracing was done manually in Amira 2019.3 using the filament editor in linear tracing mode. (…) The total filament lengths were extracted from the resulting spatial graph statistics.
For Research Use Only. Not for use in diagnostic procedures.